

Looking for CRISPR genome editing protocols? Read about our growing library of protocols and user methods—we may have what you need to get started. And, if you have a novel protocol for using Alt-R CRISPR guide RNA and/or nucleases, find out how to share it with the research community.
Research into applications using CRISPR-Cas systems for genome editing is expanding quickly. This presents a need for effective reagents and protocols for a wide variety of model systems beyond mammalian cultured cells.
CRISPR guide RNAs. Use Alt-R CRISPR guide RNAs (gRNAs) to direct potent, on-target genome editing. These length-optimized RNAs are chemically synthesized, which allows addition of modifications for increased nuclease resistance and reduced innate immune responses.
CRISPR proteins. Choose from several recombinant variants of Cas9 (Streptococcus pyogenes), as well as Cas12a (formerly Cpf1) (Acidaminococcus sp. BV3L6) endonucleases. Alt-R S.p. Cas9 Nuclease V3 is suitable for most genome editing studies. Some experiments may benefit from use of Alt-R S.p. HiFi Cas9 Nuclease, a high-fidelity nuclease that has been engineered to reduce off-target effects while retaining the on-target potency of wild-type Cas9. Alt-R Cas12a (Cpf1) nucleases provide additional genome editing target sites, because they recognize an AT-rich protospacer adjacent motif (PAM) rather than the CG-rich Cas9 PAM sequence.
If you are interested in knock-in experiments or other applications requiring homology-directed repair (HDR), consider using an Alt-R Cas9 nickase to create single-strand breaks. Use of a nickase variant with a pair of guide RNAs reduces off-target effects by requiring specific binding of 2 ribonucleoproteins (RNPs). However, you will need 2 potent CRISPR target sites at an appropriate distance from your HDR target site.
CRISPR RNPs. While Alt-R CRISPR guide RNAs are compatible with CRISPR proteins from any source (e.g., cells that stably express CRISPR protein, or CRISPR proteins expressed from DNA or mRNA constructs), we recommend delivery of CRISPR reagents as RNPs by lipofection (e.g., Figure 1), electroporation, or microinjection. Cas9 RNPs are compatible with all 3 methods; however, for Cas12a RNPs, we recommend electroporation and microinjection delivery methods.
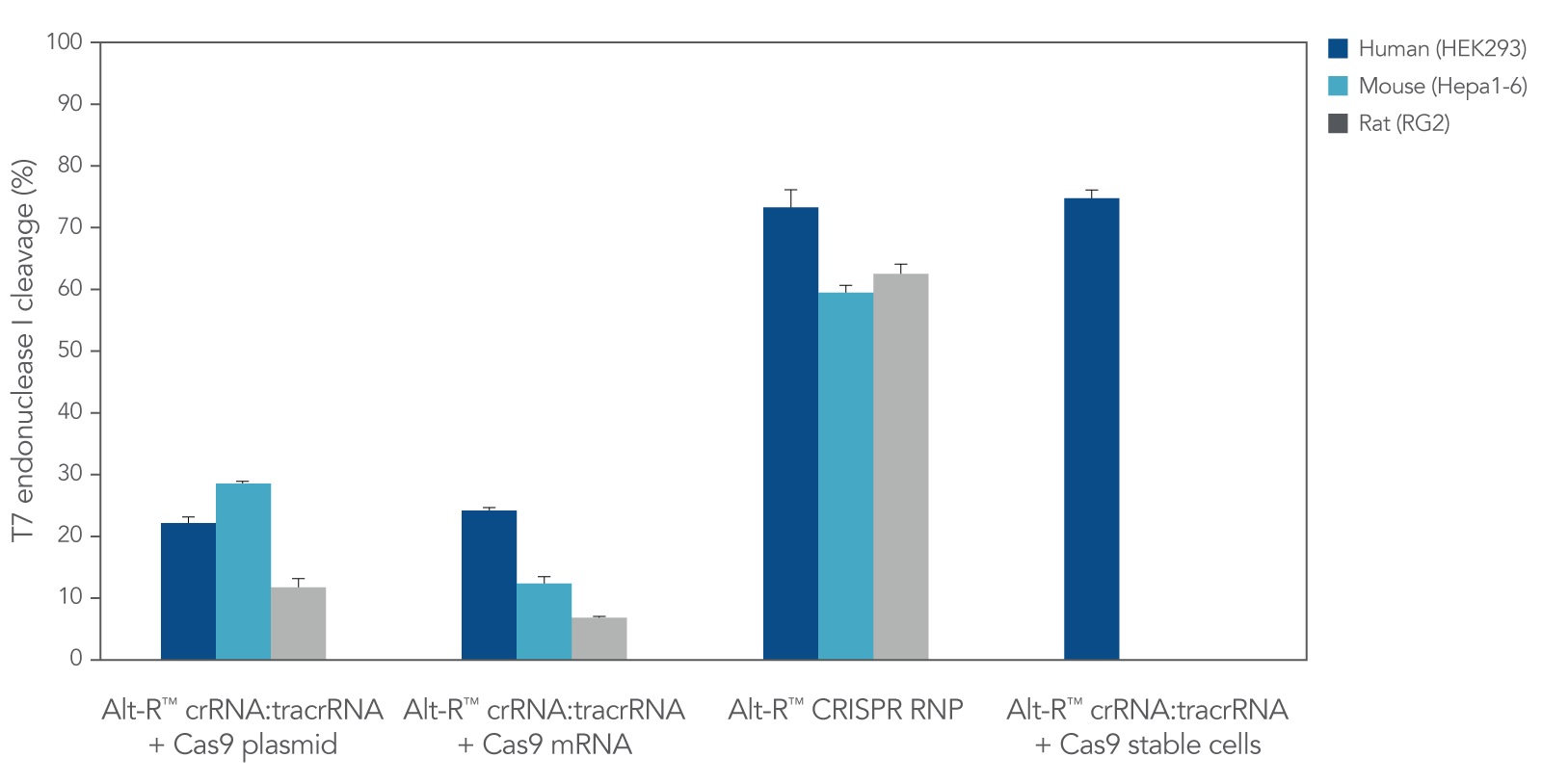
Figure 1. Lipofection of Alt-R CRISPR-Cas9 System components as a ribonucleoprotein (RNP) outperforms other transient CRISPR-Cas9 methods. Alt-R CRISPR HPRT control crRNA for human, mouse, or rat were complexed with Alt-R CRISPR tracrRNA. Resulting complexes were transfected with Cas9 expression plasmid, Cas9 mRNA, or as part of a Cas9 RNP (containing Alt-R S.p. Cas9 Nuclease 3NLS, pre-complexed with the crRNA and tracrRNA) into human (HEK-293), mouse (Hepa1-6), or rat (RG2) cell lines. The Cas9 RNP outperformed the other transient Cas9 expression methods and performed similar to reference HEK293-Cas9 cells that stably express S. pyogenes Cas9. n = 1.
IDT scientists have developed detailed lipofection and electroporation protocols for using the Alt-R CRISPR-Cas9 System and the Alt-R CRISPR-Cas12a System in mammalian cells (Table 1). With help from our collaborators, we also make user-submitted methods available for genome editing in other model systems. These protocols and methods are a good starting point for protocol optimization for your studies, and often offer tips for all aspects of genome editing experiments, from growth conditions through genome editing detection.
Table 1. Alt-R CRISPR System protocols and user methods.*
| Model system | Transfection method | IDT protocols |
| Cultured cells | Lipofection | [1] |
| Cultured cells | Electroporation | [2–5] |
| In vitro | — | [6] |
| Model system | Transfection method | User-submitted methods |
| Human pluripotent or embryonic stem cells | Electroporation | [7] |
| Mouse zygote | Electroporation | [8] |
| Mouse zygote | Microinjection | [9] |
| Zebrafish embryo | Microinjection | [10] |
| C. elegans | Injection | [11] |
* Visit www.idtdna.com/protocols to access the most up-to-date list of protocols and user-submitted methods.
We are always interested in learning about successful editing in different model systems and would like to serve as your resource for information about genome editing applications.
If you have a novel protocol for using Alt-R CRISPR RNA and/or nucleases that you would like to share with the research community, email us at CRISPR@idtdna.com to start the discussion.
Maureen Young, PhD, former Senior Scientific Writer, IDT.
Published Oct 31, 2017
Revised/updated Dec 8, 2022